IN A NUTSHELL
1. About This Work
Many Gram-negative bacteria use type VI secretion systems (T6SS) to export effector proteins into adjacent target cells. These secreted effectors (T6SEs) play vital roles in the competitive survival in bacterial populations, as well as pathogenesis of bacteria. Although various computational analyses have been previously applied to identify effectors secreted by certain bacterial species, there is no universal method available to accurately predict T6SS effector proteins from the growing tide of bacterial genome sequence data.
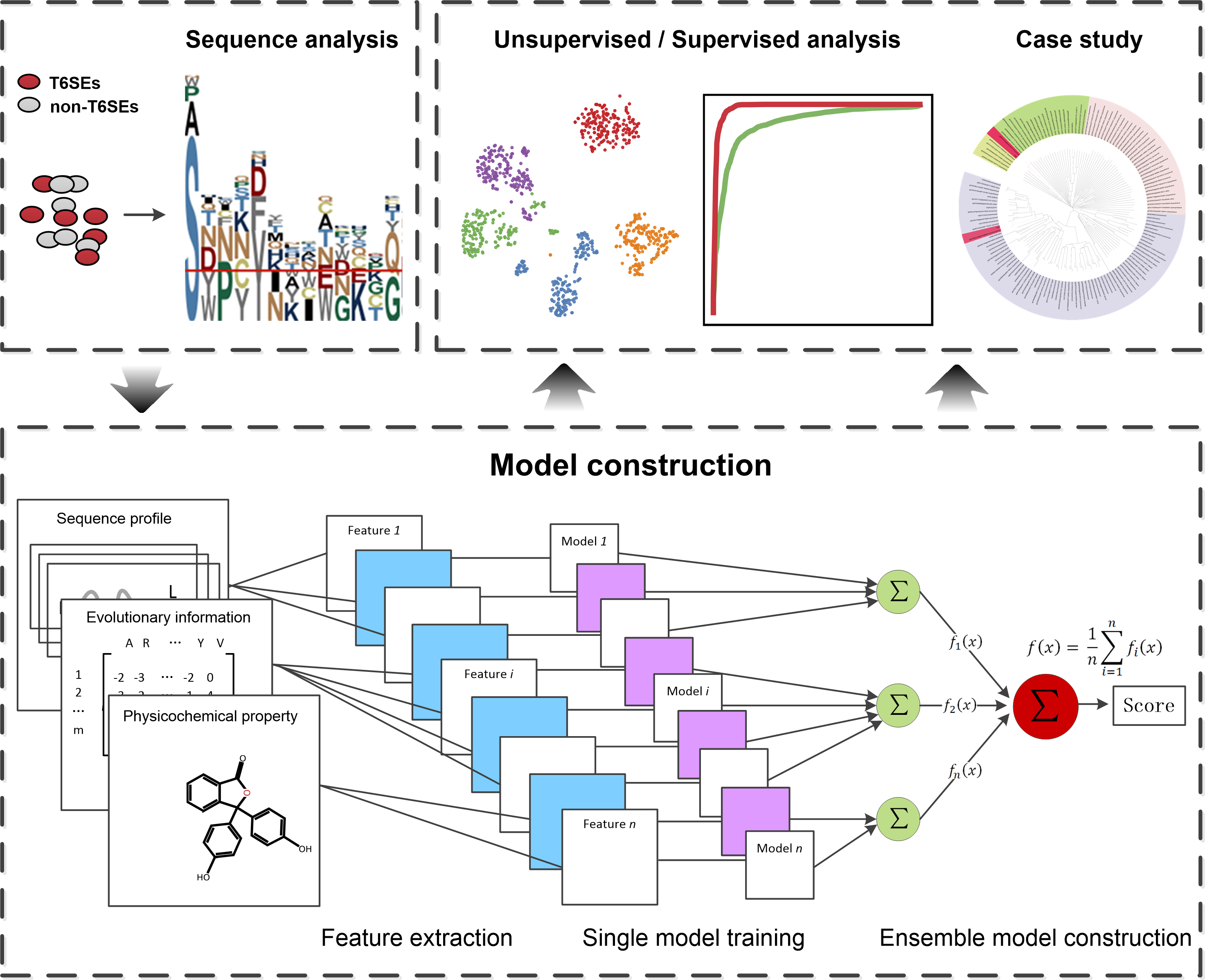
We extracted a wide range of features from T6SE protein sequences and comprehensively analyzed the prediction performance of these features through unsupervised and supervised learning. By integrating these features, we subsequently developed a two-layer SVM-based ensemble model with fine-grain optimized parameters, to identify potential T6SEs. We further validated the predictive model using an independent dataset, which showed that the proposed model achieved an impressive performance in terms of ACC (0.943), F-value (0.946), MCC (0.892) and AUC (0.976). To demonstrate applicability, we employed this method to correctly identify two very recently validated T6SE proteins, which represent challenging prediction targets because they significantly differed from previously known T6SEs in terms of their sequence similarity and cellular function. Furthermore, a genome-wide prediction across 12 bacterial species, involving in total 54,212 protein sequences, was carried out to distinguish 94 putative T6SE candidates. We envisage both this information and our publicly accessible web server will facilitate future discoveries of novel T6SEs.
2. Corresponding Authors
Trevor Lithgow, Biomedicine Discovery Institute and Department of Microbiology, Monash University, Melbourne, Victoria 3800, Australia. Tel: +61-3-9902-9217 Fax: +61-3-9905-3726 Email: Trevor.Lithgow@monash.edu
Jiangning Song, Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, Melbourne, Victoria 3800, Australia. Tel: +61-3-9902-9304 Email: Jiangning.Song@monash.edu
DATASETS
1. Construction of the Training Dataset
To construct the training dataset, we extracted 178 known T6SE sequences from the SecretEPDB database (An, et al., 2017) and 1132 non-effectors from the literature (Zou, et al., 2013), and then removed highly homologous sequences at the threshold of 90% sequence identity due to limited positive samples. We finally obtained a training dataset containing 138 positive and 1112 negative protein sequences.
2. Construction of the Independent Test Dataset
To further evaluate the performance of our proposed ensemble method, as compared with single feature based models and existing motif-based T6SE searching methods, we generated an independent dataset by extracting T6SEs from recently published works in the literature and non-T6SEs from Vibrio parahaemolyticus. After highly homologous samples (with more than 90% similarity) were removed from our training dataset, we obtained the final independent dataset with 20 positive and 200 negative samples. Aside, two very recently experimentally validated T6SEs (Lin, et al., 2017; Si, et al., 2017) were used as case studies to test the identifying capability of the proposed method.
ONLINE WEB SERVER
1. Bastion6
We developed an online bioinformatics server, termed Bastion6 (Bacterial secretion effector predictor for type VI secretion system), to provide a user-friendly T6SE prediction service. To the best of our knowledge, Bastion6 is the first machine learning based predictor for T6SE prediction. We envisage this server will be widely used to facilitate discovery of novel T6SEs.
2. Using Bastion6
Bastion6 is an online server with a user-friendly interface and few parameters, therefore it is easy to use. All you need to do is to fill the input box or upload a fasta file. The prediction job will be put into the queue system. All the jobs will be executed by Bastion6 server successively. After your job is finished, you will receive an e-mail with a url of your job result if your email address is provided.
2.1 Input Formats
Two types of input are accepted by Bastion6: sequences in FASTA format (recommended) and raw sequences.
For sequences in FASTA format, you can input as follows:
Also, the following input (which is the original formats downloaded from Uniprot database)
will be formated (without line break inside the sequence) as:
For the raw sequences, you can input as follows:
which will be formated by Bastion6 as follows:
2.2 Job run by Bastion6 ensemble predictor
A job submission will take the Bastion6 server approximately 3-5 minutes to process and accomplish the job of a typical protein sequence with 500 amino acid residues.
2.3 Input limits
1. The length of each submitted sequence should be in the range of 50 and 5000.
2. The max number of submitted sequences each time should be no more than 500.
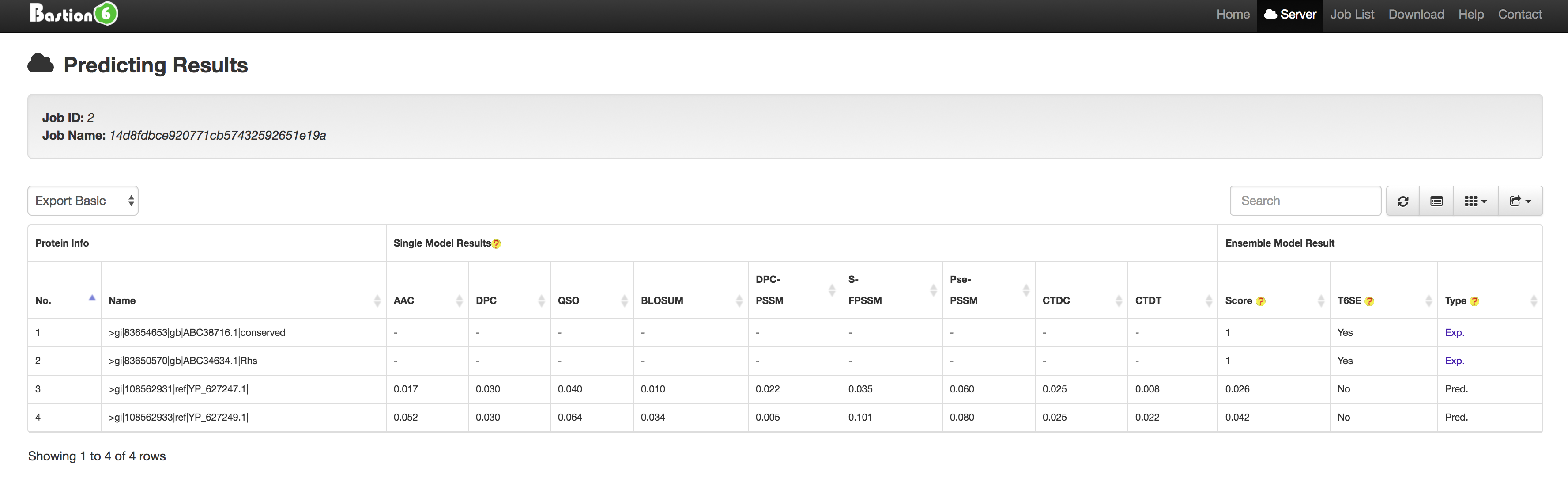
3. Bastion6 Prediction Result Instructions
There is a continuously-updated and built-in list (keeping pace with SecretEPDB) of main types of secreted effectors (such as type II, III, IV and VI secreted effectors) in Bastion6 to annotate the results after prediction, via which we aim to distinguish the known effectors from the computational predicted ones and provide detailed annotations of those known effectors for users.
For a computationally predicted protein, the result is marked as Pred. and the detailed prediction results (including single method based model results and final ensemble model result) are provided to users.
For a known type II, III, IV or VI secreted effector (such as protein Q2T2K7), the result is marked as Exp. and a corresponding url link of SecretEPDB is provided to offer detailed information for this effector (shown in the following figure).